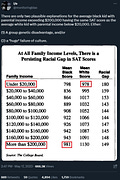
This tweet was in response to this graph posted by i/o:
Pretty easy study to do, so I figure I’ll do it here.
The NLSY97 has measurements of parent education, wealth, and income, as well as measures of child IQ (approximated with ASVAB g score, derived from 12 tests, controlled for age).
These correlate moderately with child IQ:
Parental education: .36
Maternal education: .32
(parent) Family income: .37
(parent) Family net worth: .32
Out of the 4 measures, family wealth correlates the least with child IQ, probably because some wealth is inherited, which diminishes the genetic link between the two variables. The composite of all 4 of these three variables correlates with child IQ at .45, higher than any of the 4 variables in isolation. About ~75% of this correlation is due to genetic confounding, where successful parents pass on genes for intelligence to their children (Trzaskowski, 2014).
As for whether controlling for wealth eliminates race gaps, it doesn’t. In fact, it performes reasonably worse than the composite of the 4 variables.
Table 1. Regression models predicting ASVAB g scores. *** → p < .001, ** → p < .01.
Controlling for parental wealth eliminated 14% of the difference for Blacks and 19% of the difference for Hispanics. On the other hand, controlling for parental SES eliminated 38% of the difference for Hispanics and 21% of the difference for Blacks. Regardless, these controls are unnecessary, as most of the relationship between parental SES and IQ is genetic.
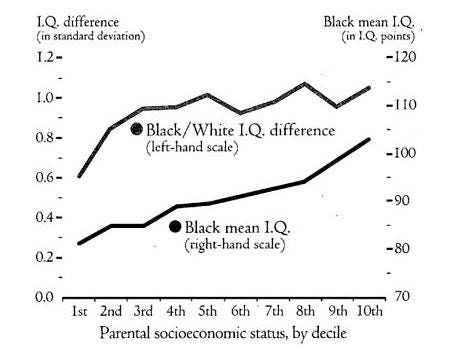
This is not a new finding. Charles Murray did the same thing in the Bell Curve, and found the same result:
Notes:
Hispanics are only White/Hispanic
Blacks/Whites are monoracial
ASVAB score is corrected for age non-linearly
Composite parental SES is done with PCA, composite ASVAB is done with FA
Wealth, ASVAB g scortes, and parental SES are standardized at mean=0 and SD=1.
R code:
new_data$white = 0
new_data$black = 0
new_data$hispanic = 0
new_data$other = 0
new_data$white[new_data$R0538700==1 & new_data$R0538600==0] <- 1
new_data$hispanic[!new_data$R0538700==4 & !new_data$R0538700==2 & new_data$R0538600==1] <- 1
new_data$black[new_data$R0538700==2 & new_data$R0538600==0] <- 1
new_data$white[is.na(new_data$R0538700) & new_data$S0191100==8 & new_data$S0191000==0 & new_data$S1224901==0] <- 1
new_data$black[is.na(new_data$R0538700) & new_data$S0191100==9 & new_data$S0191000==0 & new_data$S1224901==1] <- 1
new_data$hispanic[is.na(new_data$R0538600) & new_data$S0191000==1 & new_data$S1224901==0] <- 1
new_data$hispanic[is.na(new_data$R0538600) & new_data$S0191100==13 & new_data$S1224901==0] <- 1
for(i in 1:nrow(new_data)) {
if(!(is.na(new_data$R6889401[i]) || is.na(new_data$R6889402[i]) || is.na(new_data$R6889300[i]) || is.na(new_data$R6889405[i]))) {
if (new_data$hispanic[i]==0 & new_data$black[i]==0 & new_data$white[i]==0) {
if(new_data$R6889401[i]==1 & new_data$R6889400[i]==0 & new_data$R6889300[i]==0) {
new_data$black[i]=1
}
else if(new_data$R6889401[i]==0 & new_data$R6889300[i]==1) {
new_data$hispanic[i]=1
}
else if(new_data$R6889401[i]==0 & new_data$R6889400[i]==1 & new_data$R6889300[i]==0 & new_data$R6889405[i]==0 & new_data$R6889402[i]==0 & new_data$R6889403[i]==0) {
new_data$white[i]=1
}
}
}
}
new_data$other = 0
new_data$other[new_data$white==0 & new_data$black==0 & new_data$hispanic==0] <- 1
white <- subset(new_data, new_data$white==1)
black <- subset(new_data, new_data$black==1)
hispanic <- subset(new_data, new_data$hispanic==1)
unclaimed <- subset(new_data, new_data$hispanic==0 & new_data$black==0 & new_data$white==0)
other <- subset(new_data, new_data$other==1)
#add up exactly to 8984
wb<- subset(new_data, new_data$white==1 & new_data$black==1)
bh <- subset(new_data, new_data$hispanic==1 & new_data$black==1)
wh <- subset(new_data, new_data$hispanic==1 & new_data$white==1)
whb <- subset(new_data, new_data$hispanic==1 & new_data$white==1 & new_data$black==1)
#all 0
new_data$race = ""
new_data$race[new_data$black==1] <- "Black"
new_data$race[new_data$white==1] <- "AWhite"
new_data$race[new_data$hispanic==1] <- "Hispanic"
new_data$race[new_data$other==1] <- "Other"
new_data$Female = new_data$R0536300-1
subtestlist <- c('GS', 'AR', 'WK', 'PC', 'NO', 'CS', 'AI', 'SI', 'MK', 'MC', 'EI', 'AO')
new_data$GS = NA
new_data$AR = NA
new_data$WK = NA
new_data$PC = NA
new_data$NO = NA
new_data$CS = NA
new_data$AI = NA
new_data$SI = NA
new_data$MK = NA
new_data$MC = NA
new_data$EI = NA
new_data$AO = NA
new_data$testday = new_data$R9708601*1/12+new_data$R9708602
new_data$testday[is.na(new_data$testday)] <- 1997.661
new_data$bdate = new_data$R0536402 + new_data$R0536401*1/12
new_data$age = new_data$testday-new_data$bdate
new_data$ageat = 1997 - new_data$bdate
j = 0
for(stest in subtestlist) {
posstring = paste("R", 9705200+j*100, sep="")
negstring = paste("R", 9706400+j*100, sep="")
stcolumnindex = getcolindex(stest)
negcolumnindex = getcolindex(negstring)
poscolumnindex = getcolindex(posstring)
new_data[, stcolumnindex] = as.numeric(pmax(new_data[, negcolumnindex]*-1, new_data[, poscolumnindex], na.rm=TRUE))
new_data[, stcolumnindex] = normalise(new_data[, stcolumnindex])
new_data[, stcolumnindex][!is.na(new_data[, stcolumnindex])] <- agecorrectasvab(stest, normalizeit=T)
j = j+1
}
#head(new_data$GS)
#sd(new_data$GS, na.rm=T)
#cor.test(new_data$GS, new_data$AR)
#cor.test(new_data$WK, new_data$PC)
#cor.test(new_data$NO, new_data$CS)
#cor.test(new_data$AI, new_data$SI)
#cor.test(new_data$MK, new_data$MC)
#cor.test(new_data$EI, new_data$AO)
#cor.test(new_data$GS, new_data$age)
#mean(new_data$age)
#min(new_data$age)
#max(new_data$age)
#mean(new_data$ageat)
#min(new_data$ageat)
#max(new_data$ageat)
iq <- subset(new_data, select = c(GS, AR, WK, PC, NO, CS, AI, SI, MK, MC, EI, AO))
new_data$g2 = NA
new_data$g2 = getpc(iq, normalizeit=T, fillmissing=F, dofa=T)
wh <- subset(new_data, new_data$white==1)
new_data$g2 = (new_data$g2 - mean(wh$g2, na.rm=T))/sd(new_data$g2, na.rm=T)
#cor.test(new_data$g2, new_data$skincolor)
#cor.test(new_data$g2, new_data$GS)
#cor.test(new_data$g2, new_data$AR)
poldat <- subset(new_data, select = c(R1204500, R1204700, R1302600, R1302700))
new_data$ses = NA
new_data$ses = getpc(poldat, normalizeit=T, fillmissing=T)
#mean(new_data$ses, na.rm=TRUE)
#sd(new_data$ses, na.rm=TRUE)
#cor.test(new_data$ses, new_data$skincolor)
#cor.test(new_data$g2, new_data$ses)
cor.test(new_data$g2, new_data$ses)
cor.test(new_data$g2, new_data$R1204500)
cor.test(new_data$g2, new_data$R1204700)
cor.test(new_data$g2, new_data$R1302600)
cor.test(new_data$g2, new_data$R1302700)
new_data$wealth <- normalise(new_data$R1204700)
sd(new_data$ses)
lr <- lm(new_data$g2 ~ new_data$black + new_data$hispanic + new_data$other)
summary(lr)
lr2 <- lm(new_data$g2 ~ new_data$black + new_data$hispanic + new_data$other + new_data$wealth)
summary(lr2)
lr3 <- lm(new_data$g2 ~ new_data$black + new_data$hispanic + new_data$other + new_data$ses)
summary(lr3)
Here are the personal functions I use
normalise <- function(vector) {
vector2 = vector
vector2 = (vector - mean(vector, na.rm=T))/sd(vector, na.rm=T)
return(vector2)
}
rcalc <- function(variable="g2", dayf) {
varindex = getcolindex(variable, dayf)
probindex = getcolindex("probpred", dayf)
ctest <- cor.test(dayf[, varindex], dayf[, probindex])
effect = ctest$estimate
pvalue = ctest$p.value
return(c(effect, pvalue))
}
cohenDinter <- function(variable="g2", rac="AWhite") {
varindex = getcolindex(variable)
varindex2 = getcolindex("racesexmixer")
deeef <- subset(new_data, new_data$race==rac)
jak = deeef[, varindex]
jak2 = deeef[, varindex2]
differ = cohen.d(jak, jak2)
effect = differ$cohen.d
pvalue = differ$p
samplesize = differ$n
return(c(effect, pvalue, samplesize))
}
cohenDpever <- function(variable="g2", rac="AWhite") {
varindex = getcolindex(variable)
varindex2 = getcolindex("pever")
deeef <- subset(new_data, new_data$race==rac)
jak = deeef[, varindex]
jak2 = deeef[, varindex2]
differ = cohen.d(jak, jak2)
effect = differ$cohen.d
pvalue = differ$p
samplesize = differ$n
return(c(effect, pvalue, samplesize))
}
agecorrect <- function(vector, normalizeit=F) {
returnit = NA
formu <- paste(vector, " ~ rcs(ageat, 4)", sep="")
fit1 <- ols(data=new_data, as.formula(glue::glue(formu)))
if(normalizeit) {
returnit = normalise(fit1$residuals)
} else {
returnit = fit1$residuals
}
return(returnit)
}
agecorrectasvab <- function(vector, normalizeit=F) {
returnit = NA
formu <- paste(vector, " ~ rcs(age, 4)", sep="")
fit1 <- ols(data=new_data, as.formula(glue::glue(formu)))
if(normalizeit) {
returnit = normalise(fit1$residuals)
} else {
returnit = fit1$residuals
}
return(returnit)
}
getpc <- function(pcdata, normalizeit=F, fillmissing=F, dofa=F) {
returnit = NA
if(dofa) {
RD <- fa(pcdata, nfactors=1, rotate="none", missing=fillmissing)
} else {
RD <- pca(pcdata, nfactors=1, rotate="none", missing=fillmissing)
}
if(normalizeit) {
returnit = normalise(as.numeric(RD$scores))
} else {
returnit = as.numeric(RD$scores)
}
return(returnit)
}
relativeweightcalc <- function(heightvec, weightvec, normalizeit=F) {
returnit = NA
formu <- paste(weightvec, " ~ rcs(", heightvec, ", 4)*Female*rcs(ageat, 4)", sep="")
fit1 <- ols(data=new_data, as.formula(glue::glue(formu)))
if(normalizeit) {
returnit = normalise(fit1$residuals)
} else {
returnit = fit1$residuals
}
return(returnit)
}
getcolindex <- function(vectorname, dayf=new_data) {
thestring = paste('\\b', vectorname, '\\b', sep="")
columnindex = grep(thestring, colnames(dayf))
return(columnindex)
}
sexcorrect <- function(vector, normalizeit=F) {
returnit = NA
formu <- paste(vector, " ~ Female", sep="")
fit1 <- lm(data=new_data, formula = formu)
if(normalizeit) {
returnit = normalise(fit1$residuals)
} else {
returnit = fit1$residuals
}
return(returnit)
}
sexualcorrect <- function(vector, normalizeit=F) {
returnit = NA
formu <- paste(vector, " ~ sexpartners", sep="")
fit1 <- lm(data=new_data, formula = formu)
if(normalizeit) {
returnit = normalise(fit1$residuals)
} else {
returnit = fit1$residuals
}
return(returnit)
}
univariate <- function(columnname = 'g2', rac = "AWhite") {
deeef <- subset(new_data, new_data$race==rac)
daf <- na.omit(deeef[, c('racesexmixer', columnname)])
zx <- glm(data=daf, racesexmixer ~ ., family = binomial)
return(summary(zx))
}
univariate2 <- function(columnname = 'g2', rac = "AWhite") {
deeef <- subset(new_data, new_data$race==rac)
daf <- na.omit(deeef[, c('pinter', columnname)])
zx <- lm(data=daf, pinter ~ .,)
return(summary(zx))
}
lineargraph <- function(columnname, xminim = -2.5, xmaxim = -2.5) {
columnindex = getcolindex(columnname)
logitlabel = "default"
innerdf = new_data
innerdf$variablename = innerdf[, columnindex]
if(columnname == 'g2') {
logitlabel = "IQ"
} else if(columnname == 'ses') {
logitlabel = "Parental SES"
} else if(columnname == 'rweight') {
logitlabel = "Relative Weight"
} else if(columnname == 'height') {
logitlabel = "Height"
} else if(columnname == 'ghon') {
logitlabel = "Honesty"
} else if(columnname == 'problem') {
logitlabel = "Problematic Behaviour"
} else if(columnname == 'skincolor') {
logitlabel = "Skin Color"
}
innerdf$race[innerdf$race=="AWhite"] <- "White"
zxcvb <- lm(data=innerdf, pinter ~ variablename*race)
zxcvb %>%
ggeffect(terms = c("variablename", "race"))%>%
plot(use.theme=F) + labs (
x = logitlabel,
y = "Proportion of Dating Partners of Another Race",
title = ""
) + xlim(xminim, xmaxim)
}
logitgraph <- function(columnname, xminim = -2.5, xmaxim = -2.5) {
columnindex = getcolindex(columnname)
logitlabel = "default"
innerdf = new_data
innerdf$variablename = innerdf[, columnindex]
if(columnname == 'g2') {
logitlabel = "IQ"
} else if(columnname == 'ses') {
logitlabel = "Parental SES"
} else if(columnname == 'rweight') {
logitlabel = "Relative Weight"
} else if(columnname == 'height') {
logitlabel = "Height"
} else if(columnname == 'ghon') {
logitlabel = "Honesty"
} else if(columnname == 'problem') {
logitlabel = "Problematic Behaviour"
} else if(columnname == 'skincolor') {
logitlabel = "Skin Color"
}
innerdf$race[innerdf$race=="AWhite"] <- "White"
zxcvb <- glm(data=innerdf, racesexmixer ~ variablename*race, family = binomial)
zxcvb %>%
ggeffect(terms = c("variablename", "race"))%>%
plot(use.theme=F) + labs (
x = logitlabel,
y = "Probability First Sexual Partner is of Another Race",
title = ""
) + xlim(xminim, xmaxim)
}
logitgraph <- function(columnname, xminim = -2.5, xmaxim = -2.5) {
columnindex = getcolindex(columnname)
logitlabel = "default"
innerdf = new_data
innerdf$variablename = innerdf[, columnindex]
if(columnname == 'g2') {
logitlabel = "IQ"
} else if(columnname == 'ses') {
logitlabel = "Parental SES"
} else if(columnname == 'rweight') {
logitlabel = "Relative Weight"
} else if(columnname == 'height') {
logitlabel = "Height"
} else if(columnname == 'ghon') {
logitlabel = "Honesty"
} else if(columnname == 'problem') {
logitlabel = "Problematic Behaviour"
} else if(columnname == 'skincolor') {
logitlabel = "Skin Color"
}
innerdf$race[innerdf$race=="AWhite"] <- "White"
zxcvb <- glm(data=innerdf, racesexmixer ~ variablename*race, family = binomial)
zxcvb %>%
ggeffect(terms = c("variablename", "race"))%>%
plot(use.theme=F) + labs (
x = logitlabel,
y = "Probability First Sexual Partner is of Another Race",
title = ""
) + xlim(xminim, xmaxim)
}
logitgraphpever <- function(columnname, xminim = -2.5, xmaxim = -2.5) {
columnindex = getcolindex(columnname)
logitlabel = "default"
innerdf = new_data
innerdf$variablename = innerdf[, columnindex]
if(columnname == 'g2') {
logitlabel = "IQ"
} else if(columnname == 'ses') {
logitlabel = "Parental SES"
} else if(columnname == 'rweight') {
logitlabel = "Relative Weight"
} else if(columnname == 'height') {
logitlabel = "Height"
} else if(columnname == 'ghon') {
logitlabel = "Honesty"
} else if(columnname == 'problem') {
logitlabel = "Problematic Behaviour"
} else if(columnname == 'skincolor') {
logitlabel = "Skin Color"
}
innerdf$race[innerdf$race=="AWhite"] <- "White"
zxcvb <- glm(data=innerdf, pever ~ variablename*race, family = binomial)
zxcvb %>%
ggeffect(terms = c("variablename", "race"))%>%
plot(use.theme=F) + labs (
x = logitlabel,
y = "Probability Individual has Dated Interracially",
title = ""
) + xlim(xminim, xmaxim)
}
theme_set(theme_ggeffects(base_size = 13))
loopthrough <- function(func, racevec, varvec) {
for(trayt in varvec) {
for(rayc in racevec) {
output2 <- cohenDinter(trayt, rayc)
print("")
print(paste("Trait: ", trayt, " Race: ", rayc))
print("")
print("")
print(output2)
}
}
}These are the most important NLSY97 variables I used:
R1204500 → family household income
R1204700 → family net worth
R1302600 → father education
R1302700 → mother education
R9705200 - R9707500 → ASVAB subtest ability scores
R9708601/2 → test date
R0538700 → source of most of my racial categories
R0538600 → source of hispanic ethnicity






I expect Nicholas J. Kurian sincerely believed what he claimed. How do you think he came to express it so confidently?